Automatic Detection of Focal Cortical Dysplasia Using Deep Learning: A Multicentre Validation Study
Abstract number :
3.237
Submission category :
5. Neuro Imaging / 5A. Structural Imaging
Year :
2018
Submission ID :
502468
Source :
www.aesnet.org
Presentation date :
12/3/2018 1:55:12 PM
Published date :
Nov 5, 2018, 18:00 PM
Authors :
Ravnoor S. Gill, Neuroimaging of Epilepsy Laboratory, Montreal Neurological Institute and Hospital; Seok-Jun Hong, Neuroimaging of Epilepsy Laboratory, Montreal Neurological Institute and Hospital; Fatemeh Fadaie, Neuroimaging of Epilepsy Laboratory, Mont
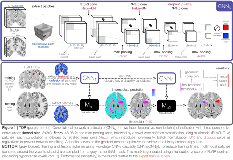
Rationale: Focal cortical dysplasia (FCD) is a surgically-amenable epileptogenic developmental malformation. Despite advances in MRI analytics, current algorithms 1-4 do not detect > 50% of FCD lesions 5. Moreover, their use requires specialized expertise, thus precluding widespread clinical application. Our purpose was to develop a novel algorithm to distinguish FCD from healthy tissue directly on MRI voxels. We propose a method harnessing feature learning capability of convolutional neural networks (CNN) 6, a powerful deep learning paradigm. Our algorithm was trained and tested on data from Montreal Neurological Institute (Site 1) and tested on independent data from S1 and 6 sites worldwide (S2-S7), for a total of 230 individuals. Methods: Volumetric T1-weighted 3D-MPRAGE (T1w) and 3D-FLAIR MRI were collected in 129 patients with histologically-verified FCD. Images underwent intensity inhomogeneity correction and standardization 7. T1w images were linearly registered to the age-appropriate MNI152 symmetric template 8. FLAIR images were linearly mapped to T1w images in MNI space.The training dataset included 40 patients (mean age: 28±9) evaluated at S1; routine MRI was initially reported as negative in 80%. This dataset served as inputs to a two-stage cascaded CNN (Figure. 1): the first CNN was designed to maximize sensitivity (i.e., detecting maximum number of lesional voxels) while the second optimized specificity (i.e., reducing false positives). At S1, a 5-fold cross-validation repeated 20 times tested sensitivity (prediction co-localizing with manual FCD labels). Specificity was assessed by testing the model on 38 healthy controls (age: 30±7) and 63 TLE-HS patients as disease controls (age: 31±8). For validation, sensitivity was tested in an independent cohort of 89 histologically-verified FCD patients (47 adults, age: 32±11; 42 children, age: 8±5) across S1 and 6 other sites. Results: For S1, sensitivity was 87±4% (average of 35/40 FCD lesions detected). In these cases, 2±1 extra-lesional clusters were detected. Specificity was 95% in healthy controls (3±1 clusters in 2/38) and 90% in TLE (1±0 in 7/63). For cross-dataset classification, overall sensitivity was 92% (82/89; 4±3 in 60). Per-site sensitivity in S1-S7 was 100% (14/14 FCD detected, 2±2), 85% (17/20, 4±2), 89% (8/9, 2±1), 75% (6/8, 2±1), 100% (8/8, 4±2), 96% (22/23, 4±4) and 100% (7/7, 2±1), respectively. Figure. 2 shows test case examples. Conclusions: We present the first deep learning multicentre study for automated FCD detection based on histologically-confirmed lesions. Operating on routine multi-contrast MRI in voxel-space, our algorithm provides the highest performance to date. We demonstrated generalizability by showing robust performance across independent cohorts with different age, scanner hardware and sequence parameters. Notably, ~50% of FCD lesions were missed by conventional MRI visual inspection. Easy implementation, minimal pre-processing, performance gains, and inference time of < 6 min/case make this classifier the ideal platform for large-scale clinical use, particularly in “MRI-negative” FCD.ReferencesAdler S et al. Neuroimage: Clin. 2017;14:18-27.Hong SJ et al. Neurology. 2014;83(1):48-55.Gill RS et al. DLMIA-MLCDS, MICCAI. 2017:349-356.Tan YL et al. Neuroimage. 2017;166:10-18.Kini LG et al. Neuroimage: Clin. 2016;11:515-529.LeCun Y et al. Nature. 2015;521(7553):436-444.Sled JG et al. IEEE Trans Med Imaging. 1998;17(1):87-97.Fonov V et al. Neuroimage. 2011;54(1):313-327. Funding: Canadian Institutes of Health Research (MOP-57840, MOP-123520)
.tmb-.png?Culture=en&sfvrsn=8a719bfe_0)