Functionalized ClinPhen: Humanized Animal Models for Detecting Pathogenicity, Interrogating Mechanism of Action, and Enabling Targeted Drug Screening in Clinical Variants
Abstract number :
2.458
Submission category :
2. Translational Research / 2D. Models
Year :
2018
Submission ID :
553547
Source :
www.aesnet.org
Presentation date :
12/2/2018 4:04:48 PM
Published date :
Nov 5, 2018, 18:00 PM
Authors :
Christopher Hopkins, Nemametrix; Kathryn McCormick, Nemametrix; Trisha Brock, Nemametrix; Gongping He, Nemametrix; and Bethan Jones, Nemametix
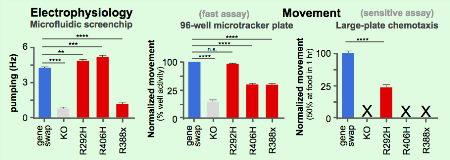
Rationale: There is growing demand for functional data on clinical variants to advance the understanding of disease mechanisms. The rapid adoption of whole genome sequencing in the clinic is generating a growing volume of data on clinical variants, much of which remains either not specified or is designated as variants of uncertain significance (VUS). Of the top 20 genes with the most submissions in ClinVar, 15.4% of variants are assigned to pathogenic/likely-pathogenic status and 8.9% are identified as benign/likely-benign. The remaining variants are either unreported (54%) or categorized as Variants of Uncertain Significance (22.1%). As a result, there is a large fraction (76%) for which better pathogenicity assessment is needed. Functional data on clinical variants is expected to be a significant contributor to pathogenicity assessment. Yet, even in cases where pathogenicity is known, clinicians and patients are often still unsure of which therapeutics offer the best treatment for a specific variant. In response to these needs, our team has created a series of humanized animal models for functional assessment of clinical phenotype ("Functionalized ClinPhen"). Methods: Animal models are created by inserting human cDNA as a gene replacement for the ortholog coding sequence in the animal. When the cDNA sequence exhibits capacity to restore normal function, the platform is ready for variant installation. Targeting epilepsy genes, we created humanized animals, each expressing STXBP1, KCNQ2, and CACNB4, at orthologous positions (unc-18, kqt-1, and ccb-1) in the C. elegans animal model. Each gene insertion was functional and high levels of rescue activity were obtained. Next, the installation of pathogenic variants in the humanized backbone yielded a series of detectable change-of-function phenotypes. Results: CRISPR-based editing was used to insert a hSTXBP1 sequence as replacement of the coding sequence of the unc-18 locus. Rescue of activity is observed at 94% by electrophysiology (up 4x from unc-18 KO), 100% in liquid "thrashing" assays for movement (up 3x), and 50% in a chemotaxis behavioral assay (up from 0%). Three known pathological variants, R292H, R406H and R388X were individually inserted into the humanized hSTXBP1 backbone and each exhibit electrophysiology and chemotaxis deficiencies. The electrophysiology assay detected both gain of function (R202H and R406H) to loss of function (R388X) alleles. Because all of the established pathogenic variants in STXBP1 are autosomal dominant, heterozygotes were examined for deficiencies in a modified chemotaxis assay. Five known pathogenic variants all showed haploinsufficiency as heterozygotes. One of the variants, p.R388X which creates a two-thirds truncation of STXBP1 protein, presented with a dominant negative phenotype as a heterozygote - approx. 3 fold more severe than a heterozygote with full gene KO. Conclusions: Drug screening is planned for all pathogenic variants of STXBP1, KCNQ2, and CACNB4 in hope of finding new or existing AEDs with likeliness to be therapeutic for the variant specific to the patient. The Functionalized ClinPhen platform is being expanded to address variant profiles in SCN1A, CACNA1A, CDKL5, ATP1A3, MAPT, TARDBP, GRN, PSEN1, APP, LMNA, POLG, MSH2 and 7000 other Rare Disease genes to help detect pathogenicity, find/repurpose drugs, and help select inclusion/exclusion criteria for patients in clinical trials. Funding: NIH grant 1 R43 AG061978-01