Harmonization of Preclinical Multicenter Plasma Protein and miRNA Biomarker Discovery in a Rat Model of Post-Traumatic Epileptogenesis
Abstract number :
1.099
Submission category :
2. Translational Research / 2C. Biomarkers
Year :
2018
Submission ID :
501375
Source :
www.aesnet.org
Presentation date :
12/1/2018 6:00:00 PM
Published date :
Nov 5, 2018, 18:00 PM
Authors :
Noora Puhakka, University of Eastern Finland; Alaa Kamnaksh, Uniformed Services University; Idrish Ali, Central Clinical School, Monash University; Gregory Smith, David Geffen School of Medicine at UCLA; Xavier Ekolle Ndode-Ekane, University of Eastern Fi
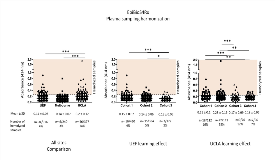
Rationale: Each year approximately 1.4 million people are diagnosed with epilepsy secondary to structural brain damage such as traumatic brain injury (TBI). There are no pharmacotherapies that can prevent epileptogenesis or mitigate the development of post-traumatic epilepsy (PTE). Unveiling biomarkers that could be used to stratify patients for antiepileptogenesis trials would increase the probability of success in therapy development and lessen the costs associated with clinical trials. Our objective was to harmonize plasma collection protocols for a multicenter-based preclinical study and to provide an objective measure for plasma quality control to identify reliable biomarkers for PTE. Methods: Epilepsy Bioinformatics Study for Antiepileptogenic Therapy (EpiBioS4Rx) includes plasma collection facilities in University of Eastern Finland (Site 1), University of Melbourne/Monash University (Site 2), and University of California, Los Angeles (Site 3). TBI was induced using a standardized protocol for lateral fluid-percussion injury; control rats underwent sham surgery under identical conditions. Blood was collected at baseline and 2, 9, and 30 days post-TBI into K2EDTA tubes (0.5 ml, 2 tubes at each time point). Plasma was separated by one-step centrifugation (1,300 g, 10 min, +4°C). Sample quality was assessed both qualitatively (visual inspection) and quantitatively for the presence of hemolysis (absorbance values at 414 nm). We also performed a pilot experiment to assess if hemolysis affects measured biomarker values using reverse phase protein microarray (RPPM). Results: As of May 2018, a total of 916, 240, and 397 plasma samples were collected at sites 1, 2 and 3, respectively. Overall, samples that were non-compliant with our acceptable quality limit for hemoglobin absorbance (A414 nm < 0.25) were 4% in site 1, 3% in site 2, and 14% in site 3. The mean (± standard deviation) absorbance value for plasma samples was 0.14 ± 0.06 in site 1, 0.14 ± 0.07 in site 2, and 0.20 ± 0.12 in site 3 (p < 0.001 for site 1 vs. 3 and site 2 vs. 3). For sites 1 and 3 we observed a learning effect over time as data from multiple animal cohorts was already available. In site 1, absorbance was 0.15 ± 0.07 in animal cohort 1, 0.14 ± 0.06 in cohort 2, and 0.13 ± 0.05 in cohort 3 (p < 0.001 for cohort 1 vs. 3; p < 0.05 for cohort 2 vs. 3). In site 3, absorbance was 0.21 ± 0.1 in animal cohort 1, 0.23 ± 0.19 in cohort 2, 0.17 ± 0.09 in cohort 3 (p < 0.001 for cohort 1 vs 3; p < 0.001 for cohort 2 vs. 3), and 0.18 ± 0.06 in cohort 4 (p < 0.01 for cohort 1 vs. 4; p < 0.01 for cohort 2 vs 4). Our pilot RPPM experiment suggests that hemolysis can interfere or skew protein biomarker values obtained from the same plasma samples. Conclusions: Our analysis of plasma sample quality shows that protocol standardization and thorough training of personnel can significantly reduce the proportion of rejected samples. Furthermore, pilot analysis of select protein biomarkers in plasma provides an objective criterion for sample rejection. Funding: Supported by NINDS Center without Walls, U54 NS100064 (EpiBioS4Rx)